Posted on September 12, 2010 by candacemw

- Lane 1: Skip Lane 2: 100bp DNA Marker Lane 3: Sample A Lane 4: Sample B Lane 5: Sample C Lane 6: Sample D
The main purpose of this lab was to amplify the purple protein coding sequence in the pGBR22 plasmid using forward and reverse primers. For the most part, the lab when according to the protocol despite two errors. One, the samples were not on ice the whole time. They were out of the ice for approximately 10 minutes which could have possibly altered my results. Also, when adding the Thermopol buffer to Sample C, the intake was very frothy and bubbly so there could have possibly been an inadequate amount of the buffer solution added to the sample. Lastly, when adding the samples into the wells of the gel, the lane that contained Sample B, did not contain as much as the other wells due to a technique error on my behalf. Although these errors were small, they could have greatly altered the outcome of the results. This was the first time that I have ever did a PCR lab, so it was very interesting. From reading background information, PCR is a scientific technique which amplifies small samples of DNA by generating a lot more copies of specific DNA sequence. The samples were definitely amplified as seen on the electrophoresis gel. This lanes consisted of the following:
Lane 1: skip
Lane 2: 100bp DNA Marker
Lane 3: Sample A
Lane 4: Sample B
Lane 5: Sample C
Lane 6: Sample D
Each sample was comprised of a specific amount of a 10X Thermopol Buffer solution, 10mM stock concentration of dNTP, a forward primer, a reverse primer, a DNA template (excluding Sample D), a DNA polymerase Tag, and Deionized water. The final concentration of these samples were approximately 25 uL. You can see the amplification of the DNA, especially in Lane 5 for Sample C. The bands on the DNA Marker corresponds to the number of base pairs of the samples. So the samples generally had anywhere from 400 to 500 base pairs.
Filed under: Uncategorized | Leave a comment »
Posted on September 12, 2010 by yilingw
This week I did DpnI treatment on my PCR reaction. This was necessary because both my accepting vector (pNIC-Bsa4) and template plasmid (pDONR221) were kanamycin resistant. pDONR221 is a bacterial plasmid so its DNA is methylated. DpnI eliminates methylated DNA so that the template plasmid from PCR isn’t carried over to transformation.
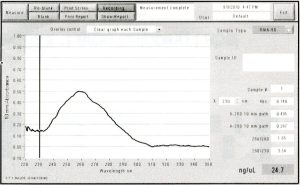
The wavelength of max absorbance was 260 nm (nucleotides absorb at 260 nm), which shows that the PCR product was conserved.
The next steps I did were cohesive end generation on PCR inserts and accepting vector. I ran into some trouble here – I did not do the two cohesive end generations simultaneously, and I left the PCR fragment and pNIC-Bsa4 in the 22 degrees Celsius water bath for too long. This, and the fact that Radhika and I spent time trying to make Kan/sucrose plates when we already had some in the fridge (they were hiding because being colirollered is very unpleasant), may have contributed to the failure of my transfomation.
The next step would be to redo cohesive end generation and annealing/transfomation. Thank goodness I made extra DNA last week!
Filed under: Week 2 update | 1 Comment »
Posted on September 12, 2010 by thaotp

Figure 1. PCR of pGBR22. T.P. 9/10/10. Lane 1: empty. Lane 2: 100bp DNA ladder. Lane 3: 1 µl of 1:1000 dilution. Lane 4: 10 µl 1:1000 dilution. Lane 5:1 µl of 1:100 dilution. Lane 6: no plasmid.
The lab did not work as the only band appearing on the gel is the DNA ladder. The protocol and the lab notebook were compared to see what might have gone wrong. It does not seem likely that there was been a pippetting error because the person who was doing the lab at the same time also did not get the expected results, so both of us would have to independently make a pippetting error. It seems more likely that something went wrong during PCR because we used the same PCR machine. I came back into the lab after 1.5 hrs and I had to wait between 5-10 minutes more for it to finish. Either the time on the PCR machine and the clock did not match up or the PCR settings were not done correctly. Another possibility is that the agarose might have been too hot when the Ethidium Bromide was put in. Next time, the settings for the PCR machine should be done more cautiously and the agarose should be allowed to cool for a few minutes longer just in case.
Filed under: Uncategorized | 4 Comments »
Posted on September 12, 2010 by thaotp

Figure 1. Agarose gel of pGBR22 cut with restriction enzymes. T.P. 9/9/10. Lane 1: 1 kb DNA ladder. Lane 2: uncut pGBR22. Lane 3: EcoRI. Lane 4: PvuII. Lane 5: EcoRI and PvuII.
The results for lane 3 and 4 (pGBR22 cut with EcoRI and with PvuII, respectively) match with the expected results which were found using the NEB Cutter on the NEB website. For lane 5, there were supposed to be three bands which meant that the restriction enzymes did not cut fully or that there was not enough plasmid to show up on the gel (since the bands on lane 5 were very light). In lane 3, there were bands lower than the expected which meant possible contamination. The lab was not perfect, but it worked somewhat. If the lab had worked perfectly and there were leftover samples, the cut plasmid could then be amplified using PCR and that could be used in recombinant DNA.
Filed under: Uncategorized | Leave a comment »
Posted on September 12, 2010 by vdsclass
For the Virtual Screen refresher – use the CB-kin_UT.sdf for the run.
It didn’t specify and some people are using the KINASet3d.sdf. Which is fine, it will just take a long time.
Filed under: Uncategorized | Leave a comment »
Posted on September 12, 2010 by damirl
Hello everyone! This week was much more progressive for me than the last. I managed to finish my Midiprep by Wednesday and nanodropped it (twice) to get the concentration – 232.0 ng/ul (I have 100 ul of total volume). I then split this pNIC-Bsa4 with CA7 into two tubes, one of which I put in the viral evolution freezer and the other into the -20 degree Celsius fridge. During Wednesday afternoon and Thursday I did the Pymol refresher in the computer lab and completed the actual protocol but I still need to complete the mini-write up associated with it (which I plan to do today). Furthermore, in regards to the virtual refresher, I have performed my first run and plan to start the second today in hopes of wrapping that up before the meeting on Tuesday. When both are completed I will send them over to Dr.B in addition to uploading the requested parts onto GoogleDocs for you all to see. By Tuesday or Wednesday I plan to submit a sample of from my Midiprep results to be sequenced at the DNA core and hope to analyze them by the end of the week. Also, near the end of the week I will be organizing/updating my lab notebook with the past week’s work and hopefully be near starting my initial virtual screen (yay!) or perhaps performing an enzyme assay on my pNIC-Bsa4 +CA7. Good luck to everyone and I hope we all have a great week ahead of us!
Filed under: Week 2 update | Leave a comment »
Posted on September 12, 2010 by zoeoc
This is how my gel looked after PCR. I don’t know why it is so dark. Honestly, I forgot to put the ethidium bromide in before I poured the gel, but I remembered as I was pouring and put it straight into the gel. (not the best idea, I know.) I thought this was the problem, but when Thao imaged her gel, it looked the same. She did everything correctly. I don’t know what could have happened to our gels. Needless to say, I need to redo my PCR sometime this week. I would have on Saturday, but I was at my grandparent’s 50th anniversary party, something I couldn’t miss even if I wanted to. They already didn’t believe that I was in lab until 8:30 Friday night and had to miss dinner. Hopefully, this week will be much better for me and my experiments. ![ZO0910PCR[1]](https://vdsclass.files.wordpress.com/2010/09/zo0910pcr1.jpg?w=468)
Filed under: Uncategorized | 5 Comments »






![ZO0910PCR[1]](https://vdsclass.files.wordpress.com/2010/09/zo0910pcr1.jpg?w=468)
You must be logged in to post a comment.