Let’s Travel all the way back to 10/22/10. After I performed RE Digest for the second time, I had an epiphany. I have a new enzyme, CA7, so the RE Enzymes used from CAII probably will not work on my new protein. Let’s Check. Yes, it does not cut in the manner it would on CAII.
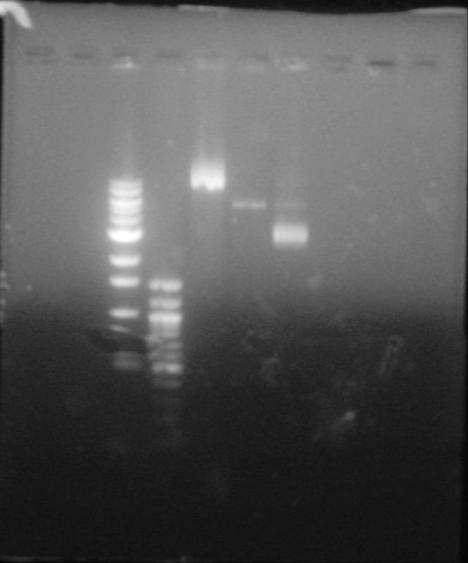
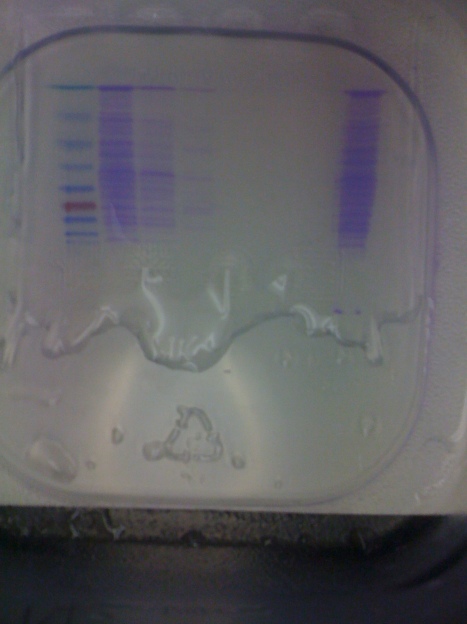
Lane I and II are skipped. Lane III and IV are the 1 kb and 100bp DNA Ladders respectively. Lane V is the uncut control plasmid of pNIC-Bsa4 with the stuffer fragment. Lane VI- is the DNA from Cloning of Colony 3 on the Master DH5alpha master plate. Lane VII- is the DNA from Cloning of Colony 7 on the Master DH5alpha master plate.
This was reassuring because I beginning to believe I was doing RE Digest improperly. Well after this, I sent off the sequence from Cloning of Colony 7 and Colony 4 to the ICMB Core, because Colony 7 resulted in four N’s for its whole sequence when the pLIC primers were used. While waiting on the results to come back, I put the DNA from Colony 3 and Colony7 in BL21 expression vectors and they came out great! I should have taken a picture of them, for everyone to see. Colony 7 looked weird again. The plate from Colony 7’s DNA from Cloning did not output distinct colonies as Colony 3 did. Now, I did add all 25 ul of the Transformation solution to the agar plate, so it could have been overcrowding in Colony 7. After this was complete, I began this week looking at the DNA sequences obtained from the core. Colony 4 came out again with CA7, but for Colony 7 from Cloning, the core gave me a sequence with 4 N’s. I am beginning to believe that Colony 7 from Cloning has the CA7 DNA in solution, but not in the pNIC-Bsa4 Vector. I choose Colony 3 to grow up my overnight culture, and then placed them in the 2 500 ml LB Broth for my OD Readings. At 600 nm, at 759 am the ABS value was .098 for B, and at 8:10 the ABS value was .110 for A. I was not able to babysit and check the OD values properly because I has class during the time of OD value check. After 2 hours, I came to check the OD values again and the 1st OD Value (ABS at 600 nm) at 11:30 was 1.606 for A and for B it was .856. (A and B come from the 2 500 ml flask that the solution of LB and bacteria were in.) After this, I added my IPTG and allowed it to incubate for 4 hours. After about 30 minutes, I realized I did not add my Kan to my new LB Solution, so I went on and added the Kan. Once that was finished, I put the LB in two tubes to allow them to centrifuge 20 min at 6000 g. The pellets weighed 48.17 grams. The pellets were resuspended and sonicated 2 days after that. Some of the solution spilled during sonication. After sonication the proteins were centrifuged again and then on to protein purification. The most tricky part of protein purification was the balancing the ph correctly because the HCL and NaOH are so concentration and a drop could totally change the pH of the protein. Question: Were my protein’s denatured? My pH went down to around 5.2 went I added the HCL, could that have been the source of the low protein yield that was to come after purification. Once the pH was at the proper value, I added the 1ml of the Ni-NTA slush to the solution and allowed it to incubate for 45 minutes. Another issues that could have arose was that I did not incubate as often as I should have to allow the protein to bind to the Nickel. Next Time, I will put the ice bucket directly in front of me and invert it every 5-10 minutes, so the Nickel will stick to the protein. I performed the rest of the task of protein purification and when the elution were taken I used the buffer that had 150 mM of NaCl instead of the 300 mM NaCl. 150 mM is close the the physiological concentration, but 300 mM creates the best protein binding environment to the Nickel. Once purification was complete, I Nano-dropped the solution to find out my protein yield. The yield for elution one was .110 at the highest. I forgot to keep the picture of the graph for that because I Nano-dropped again an kept keeping positive and negative protein concentrations. So I am back to growing up my bacteria for expression. I went into lab today to make LB and autoclave it, but the autoclave was having issues. The solutions of LB are made, but they have not been autoclaved. When I do expression the next time I have to be more careful of what I am doing and make sure I have time to babysit the LB to grow to the proper value of .6.
Filed under: GelImages, protein expression, Uncategorized | Tagged: expression | Leave a comment »











You must be logged in to post a comment.