Posted on November 10, 2010 by vdsclass
TUESDAY: Materials & Methods for Cloning is due (this will be used for your Final Report) – upload this to the Google Docs as a PDF in the
Results&Images/ReportonTarget/MaterialsandMethods Folder
Name it: UTEID_YourName_Date_ MaterialsandMethods_Cloning.pdf
Take a look at some of the research papers from Journal Club and your fellow VDSers posts for ideas of how to write it.
The final report will be in a journal format.
Filed under: cloning, Final Report, Housekeeping, Materials&Methods | Leave a comment »
Posted on October 31, 2010 by zoeoc
Good morning!!
This weekend I did a lot of virtual screening. As of right now, I have 6 libraries done, and 2 on their 1st runs. These are pretty big, so who knows when they’ll be done. I’ve glanced over the numbers, and I still don’t have ligands with higher scores than 70. I don’t know what I’m going to do about that.
Earlier this week, I miniprepped my results from the transformation. I got great numbers, but I still haven’t done the restriction enzyme digest to make sure it is the CA2 gene. Then on Wednesday, I went to the open house. I had a visitor! He was pretty knowledgeable which made me nervous, but I could handle it.
This week, I won’t be in until Wednesday (if I make it to then, that is.) On Wednesday I’m going to continue working with Dr. B on which renstriction enzymes to use. Hopefully, I can start the actual digest on Thursday. And of course, I will be working on virtual screening from my place throughout the week. I really hope I get better GOLD scores.
Filed under: cloning, Virtual Screening | Leave a comment »
Posted on October 24, 2010 by zoeoc
Since my last attempt at cloning failed, and I used up all of my PCR DNA, I had to start all over. This week I re-did PCR and the DpnI treatment (94.1 ng/ul). On Saturday I came in and did the cohesive end generation and the annealing and transformation steps. Today I made my master plate and grew up my bacteria in tubes. However, the protocol used to say 8 hours, but now it says 16, so I am hoping everything will be ok until 11 a.m. tomorrow (when I am coming in to spin down my samples). I also got the hang of virtual screening! Yay! I have been a virtual screening machine all weekend.
Top 10 Ligands from HF9 Library
1 – 70.02
2 – 68.27
199 – 68
31 – 66.39
12 – 64.37
66 – 64.16
3 – 64.10
16 – 63.96
119 – 63.95
114 – 63.93
(I feel like these are pretty low. Is that just me?)
The next steps of my research will be continuing virtual screening of course, doing the miniprep kit (hopefully Monday night), and doing the restriction enzyme digest. The miniprep kit is where I am going to stop this week. I will be MIA for a while considering I have 3 exams next week (M, T, W) and I’m leaving town Thursday after the open house. I’ll start working on the restriction enzyme stuff next Thursday, assuming nothing fails. *fingers crossed*
Have a good week everyone!
P.S. I have been keeping up with my lab notebook, and it is sooooo much easier this way. Thank you Dr. B and Kathryn for guilt tripping me into keeping up my lab notebook 🙂
Filed under: cloning, Virtual Screening | Leave a comment »
Posted on October 22, 2010 by damarcuseb

Fig. 1- 1% Agrose Gel with EtBr performed in TAE. Lane 1- 1 Kb Ladder Lane 2- 100 bp Ladder Lane 3- Cut DNA grown from Colony 1 during Cloning Lane 4- Cut DNA grown from Colony 2 during Cloning Lane 5 - Cut DNA grown from Colony 3 during Cloning Lane 6- Cut DNA grown from Colony 4 during Cloning Lane 7- Cut DNA grown from Colony 5 during Cloning Lane 8 - Cut DNA grown from Colony 6 during Cloning Lane 9- Cut DNA grown from Colony 7 during Cloning Lane 10- Cut DNA grown from Colony 8 during Cloning Lane 11- Uncut pNIC Bsa4 vector with stuffer fragment insert ******* Note All DNA was Double Digested with SspI and BsrgI Restrction Enzyme
Well this was interesting. So I tried to follow the protocol from the RE Digest that VDS performed at the begining on the year with 1.5 ug of DNA to insert into the tube so that the restriction enzymes could cut it. From that protocol 1.5 ug were added to the centrifuge tubes, but my concentration of DNA was in the 20’s (ng/ul) for all the tubes except Colony 7. This 1.5 ug was all of my DNA concentration for my samples from my Transformation I and from there I still followed the old RE Digest protocol. I added .5 ul of both Restriction Enzymes, SspI and BsrG I, 2.5 ul of NE Buffer and continued on with RE Enzyme Digest. I did the 2 hours at 37 degress Celsius then ran the gel. Once, I put the gel under UV light and looked at it, I saw one band and then it i hit me. I over-saturated my solution with DNA and did not add enough RE Enzyme to compensate for that difference. The concentration of the whole RE Solution should have been 25 ul, but mine was 60 ul due to the excess amount of DNA. Then I continued to think to myself, what else could have went wrong. Ahh! I forgot to heat block at 80 degrees Celsius for 20 minute to stop the enzymes function. Still, I sent Col 7 and Col 3 to DNA Sequencing. I got my results back from the lab, Col 7 was not a good sequence. It had a lot of N’s in the Coding Regions, but then there was hope. I apparently sent to much DNA to the sequencing lab (500 ng?), so they were going to re-run my DNA. Yes! No! I got back my results this morning and guess what it was. 5 N’s for the forward and reverse sequence. Success! Not! Well, my Colony 3 results were a little more uplifting than the Col 7 results. So I got my sequence with all the Coding Regions with DNA bases in the Coding regions and the N’s at the beginning and ends. I am excited. Then, I do a nucleotide blast and insert my sequence. As I cross my fingers hoping 7its my gene, and the results flash. It’s Carbonic Anhydrase 7. What! How did I get CA7. All the tubes I have used were labeled 40190. That’s CAII. Well, from talking to Dr. B about the matter, I would like to introduce my new enzyme CA7. Bye. Bye CAII, i do not know where you went, but CA7 is what I will be working with now. I am doing another RE Digest properly this time, and am abating my results from that. I hope that goes well, but still I forgot to cur the pNIC Bsa4 vector again. I hope the virtual gel and uncut pNIC Bsa4 control sufficient enough to give me enough for analysis.
Filed under: cloning | Tagged: RE digest | Leave a comment »
Posted on October 17, 2010 by zoeoc
Good morning VDSers-
This week I tried to clone. I grew up my (hopefully) transformed DH5alpha cells on a plate without sucrose, which is what I think the problem was. I had colonies, but they were tiny. I tried to grow them up on a master plate and in tubes overnight but nothing grew them either (which is why there are no pictures in this post). Well, ONE tube had a pellet. I am guessing that the plate was the problem because I can’t think of anything else I did wrong. Anyway, next week I have an ochem test on Wesnesday night, so I won’t be in the lab until Thursday. I am very behind on my virtual work, so next week I am going to work on that in the lab. I need to start my second run of the library we started 2 weeks ago :X I need to start running a new library, which means I have to do the clean up. Also, I need to find out which restriction enzymes to use. I will start cloning again on Friday. Being in the lab on Saturday was surprisingly nice. It was calm, quiet and no one was shoving my stuff around. I may be in the lab on Saturdays more often.
Gotta keep on keepin’ on!
P.S. Are there more skinny tube inserts for the big centrifuge? I could only spin 2 samples at a time, which was pretty inconvenient.
Filed under: cloning, Uncategorized, Virtual Screening | 2 Comments »
Posted on October 15, 2010 by damarcuseb

Fig. 1- DNA Concentration of 33.3 ng/ul and the DNA grown up for this Nanodrop was from the bacteria growth in the 15ml tubes which cut the Oxygen supply.
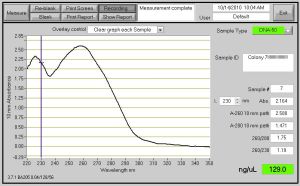
Fig. 2- DNA Concentration of 129 ng/ul and the DNA grown up for this Nanodrop was from the bacteria growth in the 14ml round bottom tubes which does not cut the Oxygen supply. Also, when spinning down the cells for this DNA, I did not take out the toothpicks.
I have 16 pictures from Nanodrop after I transformed, grew up the bacteria and Miniprepped them. The top 2 are posted, because I figured you would not want to see 16 pictures. After the Masterplate grew up, I took a tooth pick, and sequentially added each numbered bacteria colony to a numbered tube. Each Numbered tube had 5ml of LB with 50 ug/ml Kanaymycin (5 ul of Kanamycin per tube or 250 ng for all the 8 tubes.) I did not spilt up my bacteria growth and grew them up for 8 hours straight. After my bacteria grew, I took out the toothpicks and spun them down at 5000 rpm for 5 min. Next, Mini-Prep was performed on the DNA and then came the fun part. After mini-prep (I eluted in 50 ul), I proceeded to the dreadful Nanodrop. The highest DNA Concentration for the first time was 33.5 ng/ul. So, guess what, I had to perform these steps all over again. Although, I had to repeat some steps were changed along the way. When I was growing up my bacteria, I used a round bottom tube this time instead of a 15 ml colonial tube. The round bottom tubes are better because they have a loose top which enable the bacteria to consume more Oxygen for growth and the round bottom tubes are much CHEAPER than the other tubes. When spinning down the round bottom tubes, I had to parafilm the tops of them, so the liquid was secure in the tube. The first time I spun down in the 15ml tubes, I took out the toothpicks, but the second time I did not. Nothing was effected. Miniprep was done next, and the first time I eluted in 50 ul ph 8.0 water, but the second time I eluted in 30 ul of Tris-HCl. On average, my concentration of DNA from the first Miniprep was 26 ng/ul and for the second Mini-Prep it was about 52 ng/ul. In conclusion, the second time I minipreped with less elute I got a stronger concentration of DNA for use of RE Digest and DNA Sequencing. Also, for RE Digest the enzymes I will be using are SspI and BsrgI.
Filed under: cloning | Tagged: clones, colonies | Leave a comment »
Posted on October 15, 2010 by damarcuseb

Fig. 1- Image of the Master Plate after transformation was successfull and 8 colonies were hand selected to put placed in a numbered grid (1-8). When DNA is grown up, it will be easier to go back and refer to the colony that has the insert or not, because the numbered tubes help organize colony growth and cloning. The agar plate on the bottom contains the protocol mix of Cohesive Generation solution of the Accepting Vector (2 ul) and of the CAII Insert (4ul). The agar plate on the top contains my own remixed amount of accepting vector(2ul) and CAII Insert(8ul). Note: Each Agar Plate contains 5% Sucrose and Kanamycin Resistance. The Sucrose is for negative selection of colonies without the insert.
Yes! Bacteria grew up on my original plate and I had enough colonies to make a master plate. When I performed all the steps to get to the image that is shown, I had to prepare my pNIC-Bsa4 accepting vector, so that it is empty and has the chance to take up my plasmid. After that, I had to perform cohesive end generation on my CAII insert and my pNIC-Bsa4 accepting vector. I did not know what co-hesive en generation was so I looked it up. Dictionary.com gave this definition. Cohesive End – Single-strand extension on each end of a duplex DNA molecule that is usually produced by restriction endonuclease digestion and which facilitates ligation of two similarly cut DNA molecules. Called also sticky ends. Also, the t4 DNA Polymerase Buffer is the NEBuffer 2. This was found in the catalog, in the VDS Solutions cabinet. When annealing and transformation was ready to be accomplished I did two different mixes of Accepting Vector and CAII Insert, one from the protocol and another from whatever I wanted. The DH5aplha cells I used were the Invitrogen One Shot Max Efficency Tr1 cells. When all my solutions were prepared and I could spread my bacteria with the plasmid on my plate, I used the metal loop, which I heat steralized and let sit for 30 sec so it does not burn my bacteria. After this was incubated overnight, it grew up about 15 colonies, then was stored in the fridge. On Monday, October 11th, I picked 8 colonies to be put on a master plate and grew them overnight, so I could grow them up in tubes next.
Filed under: cloning | Tagged: clones, colonies | Leave a comment »
Posted on October 10, 2010 by zoeoc
Hey–
I forgot to e-mail myself the nanodrop pictures, so sorry Dr. B for more text. Last week I had to spend alot of time in the lab to make up for my week before OU. I got a lot done but still have muchhhhh more to do. I ran a gel with half of my 1st PCR samples, but I ran it off the gel backwards. BLACK TO RED cables fyi. Not to mention, I dropped that gel on the floor. Nonetheless, I had to use the last half of my samples to run another gel. The 4 mM MgCl2 was the brightest band, so I ran another PCR with that concentration in all of the tubes. It worked. The pictures are shown in my last post which I updated mid-week.
https://vdsclass.wordpress.com/2010/10/03/not-much-progress/
I started the cloning process by prepping 3 samples of pNIC-Bsa4. I am worried because Yiling said on her post that the concentration should be 50 ng/ul but mine were only 16.8, 20, and 19.3 ng/ul. I also cleaned up my PCR samples. Everything was going well until I had to put them at 80 degrees Celcius for 20 minutes. I didn’t know where the 80 was on the heat block, so I just guess-timated. Then, I went to the Health Professions Office the bathroom while I was waiting. When I came back the heat block was on 110 degrees Celcius!!!!!!!!!!!!! I thought for sure that my DNA would be denatured but I nanodropped it and miraculously, the concentration was 140.6 ng/ul. Although there is still DNA in my samples, I’m not so sure what condition it is in after being in that sort of heat, so my cloning may not work this week. I also started screening my first library on Tuesday, but the first won’t be finished until this Tuesday. I will be sure to post the results when the 2nd run is done.
I had a wedding to go to this weekend, so I am just starting my paper :'[ (And yes, I did start a rough draft last week and realized I was doing the wrong disease. I am not lazy and procrastinating. Red Bull will have to give me some major wings tonight!) And just my luck, my journal club is this week. I will start cloning on Thursday and hopefully run another library over the weekend. Wish me luck!
P.S. What is the Pymol image for the contest supposed to be of? Just our target?
Filed under: cloning, PyMol Image Contest | 6 Comments »
Posted on October 4, 2010 by vdsclass
Zoe asked:
I am coming in to make my gel and run the samples. I was looking at the protocol, and it says “Use 100kb ladder as marker” Then it says right under that 100 bp ladder. Which should we use? How much blue juice should I put in each sample (2 uL?) ? How long do we need to run the gel for and how far down should our samples be?
Thank you,
Zoe
Dr. B says:
Yes – TYPO – should be 100 bp ladder.
amount of blue juice depends on the volume of your sample.
Remember to only take HALF of you PCR sample.
So that should be 12.5 ul if you started with 25 ul.
Then BLUE JUICE is 6X concentration stock and you want it to be around 1X final in the tube..
So, 12.5 divided by 6 is roughly 2 ul.
The blue dye front of you gel should be at least 3/4 th of the way down the gel.
Dr. B
Filed under: cloning, GelImages, Protocols | Tagged: help please, PCR, Targets | Leave a comment »
Posted on October 3, 2010 by yilingw
Today, I realized I forgot to take a picture of my plates so I went to lab. The camera battery died. So here is a drawing of my plate with 8ul of CA7 PCR treated insert:

The bacteria colonies were white, but the agar wasn’t orange (don’t worry – I didn’t add any ethidium bromide). The reason the agar is orange in the picture is that I needed the agar to be a darker color so the colonies would show. These results made me happy. I thought my transformation failed last week. But when I looked at my petri dish after 3 days, I saw some definite bacteria growth. This and the fact that I added more of the PCR insert contributed to a successful transformation.
I also made a Master plate, spun down my sample tubes and Miniprep-ed them. I didn’t see any growth on my Master plate, but sample tubes 3 and 6 had bacterial growth. When making the Master plate, I used both the pipette tips and toothpicks to transfer the colonies. I think I prefer the toothpicks because I always poke into the agar when I use pipette tips. Other than working on my transformation, I also started Midiprepping the pNIC-Bsa4 with the high-speed kit.
Next step: On to restriction enzyme digest!
Filed under: cloning | 2 Comments »
Posted on September 26, 2010 by yilingw
Sunday, September 26th, 2010
2:00 p.m.
I attempted my third transformation on Friday, which reminds me – I need to check on my plates today!
2:42 p.m.
The moment of truth… dun dun dunnn.. Nothing!
I made two plates this time – one with 4 ul of T4-treated insert and the other with 8 ul. I was expecting the plate with 8 ul to grow more since it would be more likely for the accepting vector to take up the PCR insert if there was a higher concentration of inserts. Oh well, more time to work on that research report, huh?
Also, I found a coliroller in my shoe!
Filed under: cloning | 3 Comments »
Posted on September 25, 2010 by kimberlyrj
Finally some success! On Monday morning a sent in Sample 3 and 4 (both of which came from the same colony) to DNA sequencing. #3 reflected the appearance of the other samples and well as looked like cut pNIC-Bsa4 so I assumed it had failed. #4 looked slightly different but still did not look like the online simulation of the cut Zhang gene. When I got the results back, I blasted the forward and reverse sample 4 with Zhang and it came out as a perfect match. This was much better than I expected. Then, I had to blast #3 because if it turned out to not be Zhang, I would have a mixed colony which could cause problems in the future. It also matched much to my surprise. Then I found the colony on my original plate that both samples came from and grew up 2 flasks of 80ml each of this colony. It grew over night and left me with large pellets after being spun down. Next week I’m going to send a sample to DNA sequencing again because my plate had overgrown a bit and the colony might have been tainted. Hopefully I will be moving forward next week.
On a different note, if anyone can tell me how to embed images, I would really appreciate that. Hipe you all have a fun weekend!
Filed under: cloning | Tagged: clones, help please, success | Leave a comment »
Posted on September 24, 2010 by vdsclass
We have a new Midi Prep kit. Adam actually got us the Hi -Speed kit – which should be alot faster.
But, the steps are a little bit different. When you use it – be sure to use the instructions in the box.
HiSpeed Plasmid Midi Kit
http://www.qiagen.com/products/plasmid/qiagenplasmidpurificationsystem/hispeedplasmidmidikit.aspx
Filed under: cloning, Housekeeping, Protocols | Leave a comment »







You must be logged in to post a comment.