Hello VDSers!!! I hope you all had a great and restful break with good food and company. I CANNOT believe that it is the final week of class. I don’t know where the time went and I feel like this has been my toughest and fastest semester yet. But, it’s only going to get tougher and we all have to finish strong!
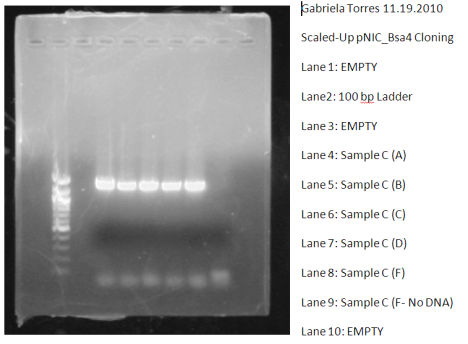
Before the lovely break, I worked on the actual pNIC-Bsa4 cloning. I got a successful PCR to work the last time from my original PCR and I chose Sample C (As mentioned in my bog post on 11/22/2010). So, since I prepared pNIC-Bsa4 as an accepting vector the day before I started cloning, I continued on topreparing the “Cohesive End Generations” for both my Accepting Vector and my PCR inserts.
For the PCR Insert I mixed in a 1.7 tube:
12ul of my PCR fragment (Sample C), 2ul of T4 DNA Polymerase Buffer(Buffer2) , 0.5ul 10X mM dCTP, 1ul of 100 DTT, 0.2ul of 100XBSA, 1ul T4 DNA Polymerase, and the rest was H2O.
For my Accepting Vector mixed in a 1.7 tube:
When I performed the PCR cleanup on my preparation of pNIC-Bsa4 as an Accepting Vector, in the final step it asks you to add 50ul of Elution Solution. Well, I accidentally added 500ul, which meant that it was diluted by a lot. So, this forced me to change the original amount of the plasmid and H2O.
39.5ul of half of cut plasmid, 5ul of 10X T4 DNA Polymerase Buffer (Buffer 2), 1.25ul of 100mM dGTP, 0.5ul 100X BSA. 1.25ul T4 DNA Polymerase, NO H2O.
These were both left at room temperature for 30 minutes (~22C) and were then heat inactivated at 75C for 20 minutes. The step that was supposed to be followed immediately after this was the annealing and transforming step, but was followed an hour after instead due to a mandatory meeting. My samples were left in the 4C while I was at my meeting.
For the annealing and tranformation step:
I made two different tubes (A & B). Tube A consisted of 2ul of T-4 treated Accepting Vector along with 4ul of each T-4 treated Inset. Tube B consisted of my own mixture and I added 2ul of the Accepting Vector and 8ul of the Insert.
They were left at room temperature for 10 minutes, moved to ice, and then 25ul of competent cells were added. They were then left on ice for another 30 minutes, heat shocked for 30seconds at 42C , put back on ice, SOC was addedand they were then left on the 37C shaker for an hour. I did not have KAN+Sucrose plates available for me, so Dr. Beckham allowed me to used KAN plates alone. I spread all of the itranformationl mixture to the plate using colirollers and they sat in the 37C overnight to grow.
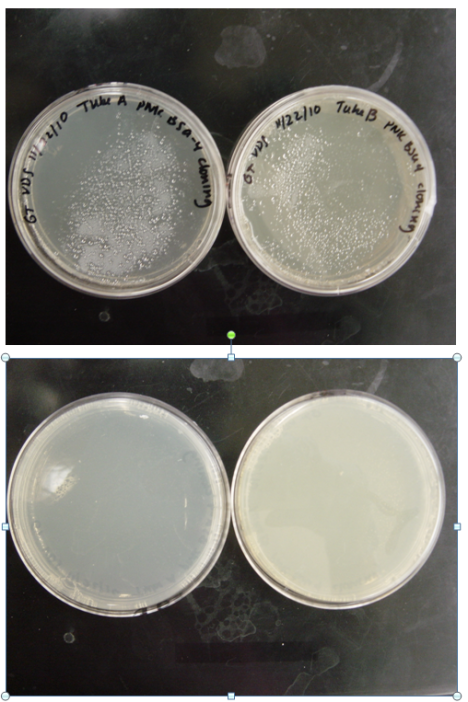
BUT, you all know how sensitive this stuff is, and it FAILED 😦 This is a picture of my plates that I took. As you can see…they have nothing grown in them!
Today, I am doing this again in hopes that I can at least have something work by Friday or Saturday. Good luck with to all of you all in lab and with your studies. The year is almost over and we have to enter 2011 with a good attitude and smile!
Filed under: Uncategorized | 2 Comments »








You must be logged in to post a comment.