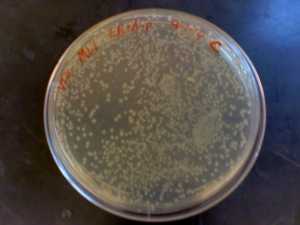
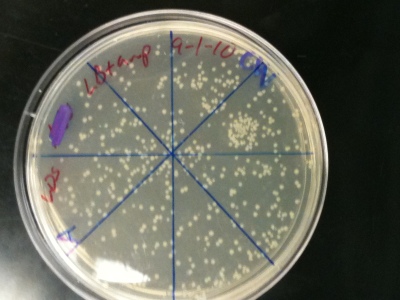
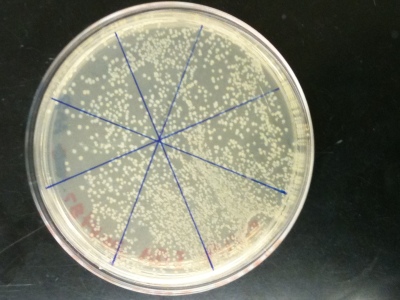
An ampicillan-resistant plasmid (pGFB) is introduced to E. coli bacteria to coax growth in its ampicillan environment. These three pictures show E. coli strain DH5 Alpha growing on LB plates treated with SOC and ampicillan. Varying amounts of plasmid (1 ng, 15 ng, and 25 ng) were added to the three plates to allow growth of the bacteria. The amount of colonies grown under optimal conditions were then counted after 16 hours. Plate C (25ng) had the highest efficiency ratio of 72.32 colonies for each nanogram of pGFB added; followed by plate A (1ng) with a 50:1 ratio and finally plate B (15 ng) with a 16:1 efficiency ratio. The plate containing the largest amount of plasmid- transformed cells was also the most efficient. The second highest efficiency ratio belonged to the plate with the lowest amount of plasmid added (A), breaking the trend. One explanation for these inconclusive results may be that the solutions of plasmid and bacteria prior to plating were not mixed properly. A large amount of a cold substance may hinder the ability of equal dispersal. The plasmid+bacteria of Plate A/C may have received slight but influential handling that led to more bacteria being transformed by the plasmid. This experiment can be performed again with a goal being to affect all of the sensitive biological material in the same manner physically.
Filed under: Uncategorized, Week1 update | 2 Comments »













You must be logged in to post a comment.