VDS Staff,
The notebook for this one is due until on FRIDAY (unless they do the lab on Sunday – then it is due Sunday night)
No lab report due for this one until after the Next wet lab (Characterization) – all they have to do is Post their Images from the lab to the Wikispaces.
There are 2 parts to the lab
PART 1: Purification
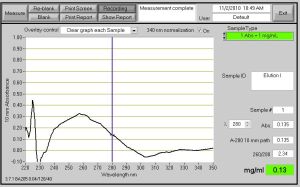
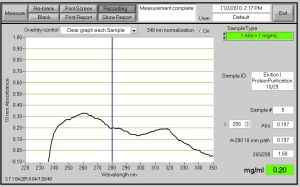
PART2: Nanodrop to measure amount of protein (this does not have to be done at the same time – but if the Nanodrop is available, they should do it)
The first steps are incubation steps that take awhile (so – to save time – they should immediately start these and then fill in their lab notebook and do Pre-lab calculations)
– while they are waiting they can 1. do pre lab calculations 2. download the Journal Paper and look up the Maximum Wavelength for the last part of the lab
REAGENTS:
Don’t let them use TOO much Ni-NTA resin – it is expensive.
Don’t let them set the Benzonase out too long (must keep on ice)
For the Flow Through and Wash steps – they can use the ‘really cheap’, non-sterile tubes that we used for the Beer’s law lab. They should tape to label these – since they can be washed and re-used.
However, for the Elution 1 and 2 – they should used ‘fancy’ purple screw cap tubes
IMPORTANT CONCEPTS & SKILLS:
Emphasize keeping the protein and re-agents cold because the protein could denature.
What the lysozyme does
What the Benzonase does
Our protein is soluble and is found in the soluble fraction after clarifying the lysate
(from the cytosol – not a membrane bound protein)
The HIS tag on our protein binds to the Nickel on the beads (resin = beads = matrix)
The HIS tag is not ‘natural’ – it was put on their by genetic modification
What imidazole does. It competes with the Histidines of the HIS tag for the binding with Nickel. If you add enough imidazole – the protein will come off the beads.
TROUBLESHOOTING:
If there Elution 1 doesn’t come out purple – they should re-incubate their Flow Through and Wash with the Ni-NTA and let it run through again.
Filed under: Mentors, protein expression | Leave a comment »



You must be logged in to post a comment.