Posted on September 13, 2010 by mikejw1
This week (in review) I was able to do the restriction enzyme digest and pcr protocols. My restriction enzyme digest came out successful in that all the bands of broken down pGBR22 plasmid were visible, but not the right number. Possible causes most likely deal with the amount of added restriction enzymes in relation to the plasmid. Also, the bands were bright, indicating a gratuitous amount of plasmid present. To conserve lab resources the amount used will be better monitored during future exercises. The PCR protocol calls for another look and more poignant labeling this following week (a mix-up at the incubator led to an a few extra hours of making another pvuII sample). The first gel (5 ul of a,b,c, & d each) and second (20 ul of each) did not yield any results. My reasoning for doing the 5 ul first was that it would take less time, which Dr. B and a clock assured me later that it did not. Again, I believe the handling of the time and temperature sensitive enzymes is to blame. 100 bp ladders were the only thing to show up on the gels when viewed under UV. I’ll be redoing this lab again to achieve readable results. Another week, another lab myth BUSTED!
Filed under: Uncategorized, Week 2 update | Leave a comment »
Posted on September 12, 2010 by yilingw
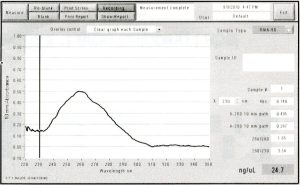
This week I did DpnI treatment on my PCR reaction. This was necessary because both my accepting vector (pNIC-Bsa4) and template plasmid (pDONR221) were kanamycin resistant. pDONR221 is a bacterial plasmid so its DNA is methylated. DpnI eliminates methylated DNA so that the template plasmid from PCR isn’t carried over to transformation.
The wavelength of max absorbance was 260 nm (nucleotides absorb at 260 nm), which shows that the PCR product was conserved.
The next steps I did were cohesive end generation on PCR inserts and accepting vector. I ran into some trouble here – I did not do the two cohesive end generations simultaneously, and I left the PCR fragment and pNIC-Bsa4 in the 22 degrees Celsius water bath for too long. This, and the fact that Radhika and I spent time trying to make Kan/sucrose plates when we already had some in the fridge (they were hiding because being colirollered is very unpleasant), may have contributed to the failure of my transfomation.
The next step would be to redo cohesive end generation and annealing/transfomation. Thank goodness I made extra DNA last week!
Filed under: Week 2 update | 1 Comment »
Posted on September 12, 2010 by damirl
Hello everyone! This week was much more progressive for me than the last. I managed to finish my Midiprep by Wednesday and nanodropped it (twice) to get the concentration – 232.0 ng/ul (I have 100 ul of total volume). I then split this pNIC-Bsa4 with CA7 into two tubes, one of which I put in the viral evolution freezer and the other into the -20 degree Celsius fridge. During Wednesday afternoon and Thursday I did the Pymol refresher in the computer lab and completed the actual protocol but I still need to complete the mini-write up associated with it (which I plan to do today). Furthermore, in regards to the virtual refresher, I have performed my first run and plan to start the second today in hopes of wrapping that up before the meeting on Tuesday. When both are completed I will send them over to Dr.B in addition to uploading the requested parts onto GoogleDocs for you all to see. By Tuesday or Wednesday I plan to submit a sample of from my Midiprep results to be sequenced at the DNA core and hope to analyze them by the end of the week. Also, near the end of the week I will be organizing/updating my lab notebook with the past week’s work and hopefully be near starting my initial virtual screen (yay!) or perhaps performing an enzyme assay on my pNIC-Bsa4 +CA7. Good luck to everyone and I hope we all have a great week ahead of us!
Filed under: Week 2 update | Leave a comment »
Posted on September 11, 2010 by candacemw

Lane 1: skip Lane 2: 1kb DNA Ladder Lane 3: Uncut Plasmid Lane 4: PvuII Lane 5: EcoRI Lane 6: PvuII + EcoRI
The restriction enzyme lab was very interesting and I learned a lot from this process. Restriction enzymes are enzymes that can cut DNA at specific points on the strand. The restriction enzymes used were PvuII and EcoRI. EcoRI incubates at 37 degrees Celsius and denatures at 65 degrees Celsius for 20 minutes and works with NEBuffer 1,2,3,4. PvuII incubates at 37 degrees Celsius and does not denature at any temperature and also works with NEBuffer 1,2,3,4. The lanes where comprised of:
Lane 1: skip
Lane 2: 1kb DNA Ladder
Lane 3: Uncut plasmid
Lane 4: PvuII
Lane 5: EcoRI
Lane 6: PvuII + EcoRI
In Lane 2, the PvuII has two distinct bands and in the Lane 3 the EcoRI has 1 distinct band. The bands do appear as expected and truly reflect the nature of the restriction enzymes. These bands on the gel electrophoresis are similar to the nature in which the restriction enzyme acts. EcoRI only has one band which still allows the plasmid to stay somewhat intack and heavier; that is why it pairs up with the uncut plasmid. Unlike EcoRI, PvuII makes two cuts and can be seen based on the lines made in the electrophoresis.
Filed under: GelImages, Week 2 update | 7 Comments »
Posted on September 10, 2010 by cwm733
This week I went over the 96-well plates of T107 and T152 to find hits for optimization. In particular, with T107, there were decent crystals on HT Index D2 and E6 and PegIon F9 and D1. On T152, there were decent crystals on PegIon A10. Examples of crystals are shown in Figure 1-3.
 Figure 1: T107 PegIon F9 w/ higher protein concentration.
Figure 1: T107 PegIon F9 w/ higher protein concentration.
 Figure 2: T107 PegIon F9 w/ lower protein concentration.
Figure 2: T107 PegIon F9 w/ lower protein concentration.
 Figure 3: T152 PegIon A10.
Figure 3: T152 PegIon A10.
I also set up 24-well optimization trays of Y107 in PEG 3350 / Tacsimate and T107 in PEG 3350 / Mg Formate at two different protein concentrations. Finally, I set up a 24-well optimization tray of T107 in L-Proline, HEPES, and PEG 3350 (HT Index F1).
I also learned to use DSF assay and ran an assay of DMSO (control) vs. BAS (gold standard) vs. 14-3-187 with NS1A-ED wild type, Y107, A107, T107, and T152.
Filed under: Week 2 update | Leave a comment »







You must be logged in to post a comment.